Accurate anatomical evaluation of the gastrointestinal tract is essential for clinical applications such as fistula localization, lesion mapping, and radiotherapy planning. A bowel topological skeleton provides an interpretable centerline-like representation of the intestinal trajectory, enabling regional annotation, shape registration, and clinically meaningful localization along the bowel path. However, small bowel anatomy is exceptionally challenging: it is long (up to ~6 m), tightly coiled, highly variable across subjects, and frequently exhibits self-contact and deformation, making complete 3D skeleton tracing extremely time-consuming and error-prone for clinicians.
While deep learning has achieved strong performance for small bowel segmentation, direct skeleton extraction remains difficult. Existing approaches often depend on manual skeleton labels and are vulnerable to topological failures in self-contact or low-contrast regions, leading to fragmented tracks, spurious loops, or incomplete paths. Moreover, skeletons (sparse 3D curves) and masks (dense volumetric regions) differ substantially in representation and feature distributions, complicating unified modeling.
Key Requirements
- Preserve a single continuous, anatomically valid path
- Reduce dependence on manual skeleton annotations
- Remain computationally efficient for large 3D volumes
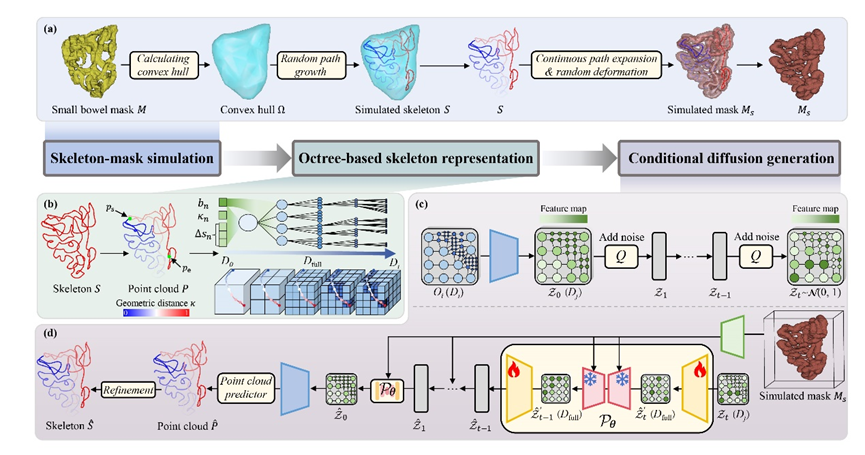
We propose Tree-Diffusion, an octree-based conditional diffusion framework that generates anatomically plausible and directionally consistent small-bowel skeletons from segmentation masks.
Core Innovations
- Topology-simulation pipeline: We synthesize anatomically consistent skeleton-mask pairs as geometric priors, substantially reducing reliance on manual skeleton labeling.
- Octree latent representation with cross-domain alignment: A compact latent space via a VAE with hierarchical octree sparse encoding captures sparse geometry and global direction; octree-domain and image-domain modules jointly preserve local directionality and global shape consistency.
Across four large-scale abdominal CT datasets, Tree-Diffusion achieves superior topological completeness, geometric accuracy, and generalization compared with prior skeletonization pipelines, while remaining efficient through sparse octree modeling.